DNA sequencing has helped develop various sectors of the economy and mainly in the health sector. DNA sequencing has helped scientists understand the causes of related genetic disorders and how to cure them. DNA sequencing has also helped scientists understand the evolution of different animal species. What about developing GMOs? Yes, DNA sequencing plays a vital role in developing genetically modified animals with certain targeted features and a high production level.
What Is DNA?
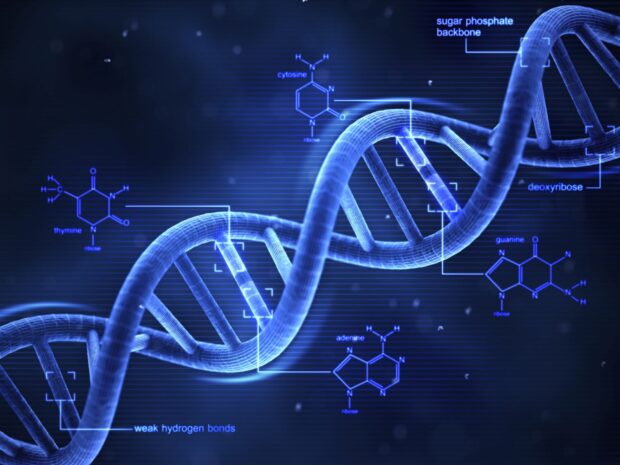
DNA molecule, also known as deoxyribonucleic acid, carries genetic instructions and information that cells require for development, proper functioning, reproduction, and growth.
The structure of DNA consists of twisted double strands of nucleotides joined together in a chain by hydrogen bonds.
What Is DNA Sequencing?
DNA sequencing is a scientific process that determines the sequence of the basic units of DNA molecules, the nucleic acids. The sequence of the nucleic acids provides information on the arrangement of nucleotides bases in a fragment of DNA strand. For comprehensive information on DNA sequencing.
Nucleotides bases found in DNA are; Adenine(A), Cytosine(C), Guanine(G), and Thymine(T).
History of DNA Sequencing
The order of nucleic acids in DNA strands finds its importance in many different fields of research. However, the extent of DNA sequencing depends on the technology in use. Due to the changes and advancements in modern technology, the techniques of sequencing have also evolved. Advanced tech has made it possible to sequence a whole genome instead of a short nucleotide.
Below is a series of DNA sequencing generations taking you through the changes that have taken place over the years.
In 1953 Watson and Crick produced the first 3-D structure of DNA. However, the ability to sequence the DNA came later in 1977, when the first DNA sequencing technology came into light.
1. First DNA Sequencing Technology
The first DNA sequencing was in the 1970s with different technologies;
- The Maxam-Gilbert method
- The Sanger’s chain termination technique or the dideoxy technique
Sanger’s method became famousfor the two and involved synthesizing the DNA chains on a template strand. However, the process had drawbacks in that the chain growth would stop once a nucleotide that lacks 3’ OH, hydroxyl group joined the chain. The process produced truncated DNA molecules. The process then involved separating the molecules according to size and charge through electrophoresis, and a computer then decoded the DNA sequence. Click here for more information on DNA sequencing technology.
However, later the method would change, and automatic sequencing machines would separate the truncated DNA molecules by size within glass capillaries. Then lasers would detect the DNA molecules through the laser excitation process.
2. Second Generation Sequencing

The second generation of DNA sequencing technology has dramatically surmounted the first-generation DNA sequencing technology. The secondgeneration has set a platform for the nextgeneration of DNA sequencing.Pal Nyren and his colleagues pioneered this process, known as pyrosequencing. Pyrosequencing involved the use of the luminescent method to measure the rate of pyrophosphate synthesis. The process converted pyrophosphate to ATP, which acted as a luciferase substrate. In the process, the reaction produced light proportional to the amount of pyrophosphate.
The pyrosequencing method employed sequence by synthesis technique(SBS) as it required action of DNA polymerase to produce an observable output; in this case, light.
Advantages of Pyrosequencing Techniques
- The technique used natural nucleotides instead of the heavily modified nucleoside molecules
- The results occur in real-time, unlike in the dideoxy technique that required long electrophoresis.
Setback of Pyrosequencing
One major setback of pyrosequencing was in determining the number of identical nucleotides in a row. The amount of light signified the length of the homopolymer,but it was still challenging to decide on the actual number of identical nucleotides in a row. However, a sound indicated a count of four or five identical nucleotides.
The pioneers later transferred the pyrosequencing technique to 454 Life Sciences company. Under 454 Life Sciences, pyrosequencing developed into a next-generation sequencing technology. The 454 pyrosequencing machines allowed parallel mass sequencing the thus increasing the number of DNA the process can sequence in a single run.
3. Use of Emulsion EPR (em-EPR)
Involves dilution and use of compartments whereby each DNA moleculeare attached to beads and then undergoes water-in-oil emulsification PCR. Each DNA molecule binds to one bead and amplifies in its droplet in the emulsion. The DNA clonally coated beads pass through the picoliter plate, and each bead fits on its washing- well.The process then involves washing the bead-linked enzymes over the picoliter plate. The charged couple device sensors below the wells then measure the amount of pyrophosphate.The amount of pyrophosphate helps determine the sequencing of the molecules.
Parallelization allowed researchers to sequence a whole single genome faster and at a cheaper rate.
Improvement of microfabrication and high-resolution imaging is what made second-generation DNA sequencing a success.
4. Third Generation Sequencing
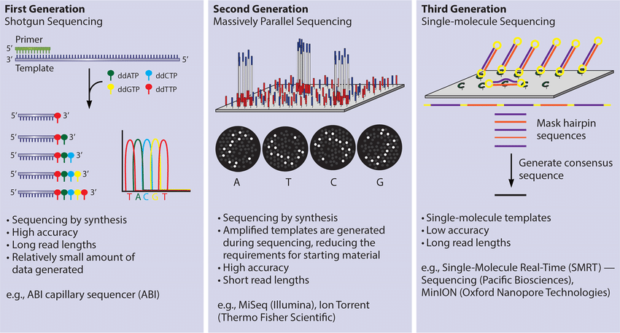
The polymerization of microfabricated nanostructures is called zero-mode waveguides(ZMWs). The polymerization and utilizes the SMRT platform.Laser excitation allows visualization of single fluorophore molecules to the bottom of the ZMWs apertures. This process can sequence single DNA molecules in a short amount of time.
PacBio machines help detect modified bases by producing kinetic energy. PacBio also has long reads, which are necessary for genome assemblies.
5. Oxford Nano Technologies(ONT)
ONT has created exciting nanoplatforms, the GridION and MinION. The MinION is a small USB sequencer generally sold to the end-users with a long read, and it’s faster and cheaper.Nanotechnology provides an opportunity to decentralize sequencing away from the usual environment. The use of nanosequencers is revolutionizing the way of doing sequencing and by who. With nanotechnology in sequencing, portability is high; hence sequencing can be done from anywhere.
Conclusion
The journey of DNA sequencing has been long, but it’s worth the pain. The traditional methods were equally important as they gave insight and a window for modern technology. From the sequencing of single DNA molecules to sequencing of paralleled DNA genomes is an outstanding achievement. The incorporation of nanotechnology in DNA sequencing has revolutionized how DNA sequencing occurs, affecting even where one can apply this technology. The confinement of DNA sequencing into specific working areas may soon come to an end. With DNA sequencing, the world can understand better how life forms.
 Comeau Computing Tech Magazine 2024
Comeau Computing Tech Magazine 2024

